Multiscale
Image Generation
The Multiscale Image Generation module offers an interface with various parameters for manipulating and configuring the MPSlib library. MPSlib features a set of algorithms based on multiple point statistical (MPS) models inferred from a training image. Currently, only the Generalized ENESIM algorithm with direct sampling (DS) mode is available.
Panels and their use
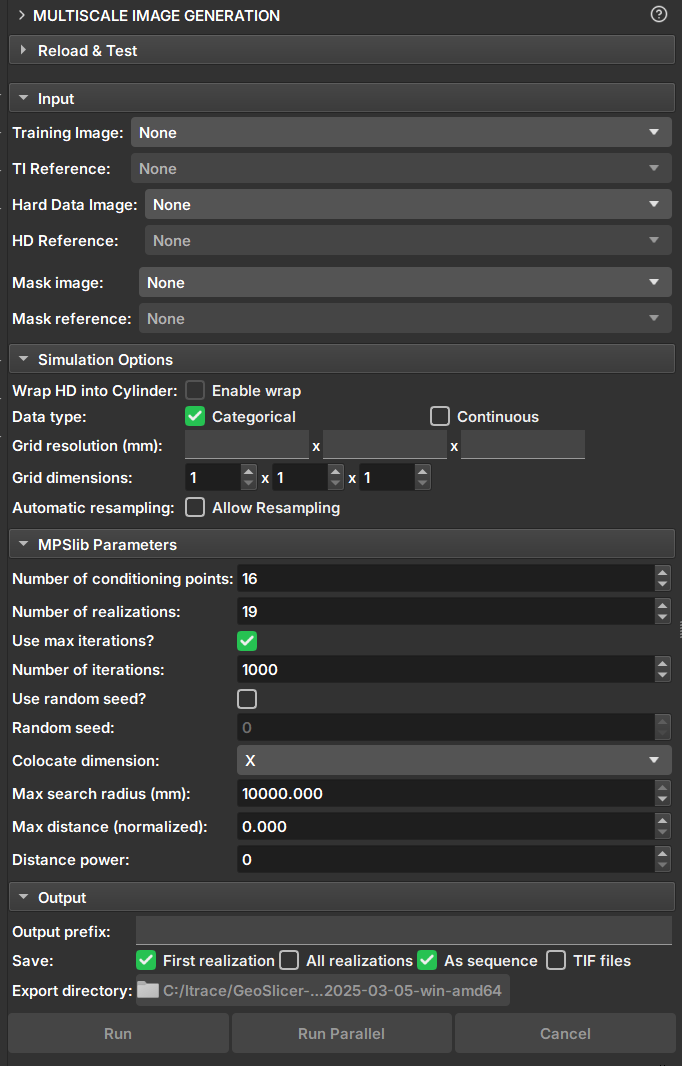 |
|---|
| Figure 1: Multiscale Image Generation Module. |
Input Data
-
Training Image: Volume that acts as a training image.
-
Hard Data Image: Volume that acts as "Hard Data", where values and points are fixed during the simulation.
-
Mask Image: Volume that acts as a mask for selecting the simulation area. Unselected segments will not be included in the simulation.
Simulation Options
-
Wrap HD into cylinder: If the "Hard Data" is a well image (2D), this option causes the image to be considered a cylinder and performs simulations as 3D.
-
Data type: Determines whether the data type is continuous or categorical. Segmentations and Labelmaps can be used for discrete and continuous simulations, but scalar volumes can only be used for continuous.
- Categorical: Segmentations control regions and determine the class value of the Hard Data and Training Image volume. Unselected segments will be disregarded.
-
Continuous: Segmentations control which regions and volume values will be used as Hard Data or training data. Unselected segments will be disregarded.
-
Grid Resolution: Voxel resolution of the resulting image (mm).
-
Grid Dimensions: Dimensions of the resulting image.
-
Automatic resampling: Activates the automatic resizing functionality of the input data to the simulation grid's dimension and resolution. If the image is an imagelog, Y-axis resizing is disabled.
- Spacing: New axis resolution after resizing.
- Size: New axis dimension after resizing.
- Ratio: Ratio of the new voxel resolution to the initial resolution.
Parameters
-
Number of Conditioning points: Number of conditioning points to be used in each iteration.
-
Number of realizations: Number of simulations and images to be generated.
-
Number of iterations: Maximum number of iterations when searching the training image.
-
Random seed: Initial value used to start the simulation. The same seed with the same parameters always generates the same result.
-
Colocate dimensions: For a 3D simulation, ensure that the order in the last dimensions is important, allowing a 2D co-simulation with conditional data in the third dimension.
-
Max search radius: Only conditional data within a defined radius is used as conditioning data.
-
Max distance (normalized): The maximum distance that will lead to the acceptance of a conditional model match. If the value is 0, a perfect match will be sought.
-
Distance power: Weights the conditioning data based on the distance from the central values. A value of 0 configures no weighting.
Output Options
-
Output prefix: Name of the generated volume or file. In case of multiple realizations, a number is added to indicate the realization.
-
Save: Options for saving the results.
- First realization: Saves only the first realization as an individual volume.
- All realizations: Saves all realizations as individual volumes.
- As sequence: Saves the realizations in a sequence set. The "_proxy" output volume indicates it is a sequence and has the controllers for visualizing the realizations.
-
TIF files: Saves all realizations as tiff files.
-
Export directory: Directory where the tiff files will be saved. It is only enabled if the "TIF files" option is selected.
Buttons
- Run: Executes the mps sequentially. The Geoslicer interface is locked during the execution of this option.
- Run Parallel: Executes the mps in parallel. In this option, the execution runs in another thread, and the interface can be used during execution.
- Cancel: Interrupts the simulation execution. Only when executed in parallel.
Post Processing
Module for data extraction after Multiscale simulation.
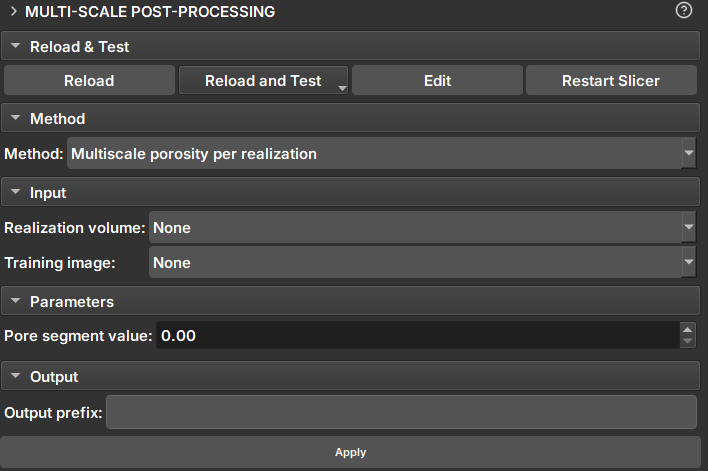 |
|---|
| Figure 1: Multiscale Post Processing Module. |
Methods
Porosity per Realization
Produces a table with the porosity percentage of each slice of a volume, across all volumes in a sequence.
Input Data and Parameters
- Result Volume: Volume for porosity calculation. If the volume is a proxy for a sequence of volumes, porosity will be calculated for all realizations.
- Training Image: Extra volume included in the calculations and added to the table as a reference.
- Pore segment Value: Value to be considered as a pore in scalar volumes (continuous data)
- Pore segment: Segment to be considered as a pore in Labelmaps (discrete data).
Pore Size distribution
Recalculates the distribution of pore size for frequency.
Input Data and Parameters
- PSD Sequence Table: Table or proxy of sequence of tables resulting from the Microtom module.
- PSD TI Table: Table result from microtom for the training image.